Publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2025
- MICCAI
 Dynamic Robot-Assisted Surgery with Hierarchical Class-Incremental Semantic SegmentationJulia Hindel, Ema Mekic, Enamundram Naga Karthik, and 4 more authors2025
Dynamic Robot-Assisted Surgery with Hierarchical Class-Incremental Semantic SegmentationJulia Hindel, Ema Mekic, Enamundram Naga Karthik, and 4 more authors2025Robot-assisted surgeries rely on accurate and real-time scene understanding to safely guide surgical instruments. However, segmentation models trained on static datasets face key limitations when deployed in these dynamic and evolving surgical environments. Class-incremental semantic segmentation (CISS) allows models to continually adapt to new classes while avoiding catastrophic forgetting of prior knowledge, without training on previous data. In this work, we build upon the recently introduced Taxonomy-Oriented Poincaré-regularized Incremental Class Segmentation (TOPICS) approach and propose an enhanced variant, termed TOPICS+, specifically tailored for robust segmentation of surgical scenes. Concretely, we incorporate the Dice loss into the hierarchical loss formulation to handle strong class imbalances, introduce hierarchical pseudo-labeling, and design tailored label taxonomies for robotic surgery environments. We also propose six novel CISS benchmarks designed for robotic surgery environments including multiple incremental steps and several semantic categories to emulate realistic class-incremental settings in surgical environments. In addition, we introduce a refined set of labels with more than 144 classes on the Syn-Mediverse synthetic dataset, hosted online as an evaluation benchmark. We make the code and trained models publicly available.
@misc{hindel2025dynamic, title = {Dynamic Robot-Assisted Surgery with Hierarchical Class-Incremental Semantic Segmentation}, author = {Hindel, Julia and Mekic, Ema and Karthik, Enamundram Naga and Mohan, Rohit and Cattaneo, Daniele and Kalweit, Maria and Valada, Abhinav}, year = {2025}, eprint = {2508.01713}, archiveprefix = {arXiv}, primaryclass = {cs.CV}, url = {https://arxiv.org/abs/2508.01713}, journal = {MICCAI}, } - arXiv
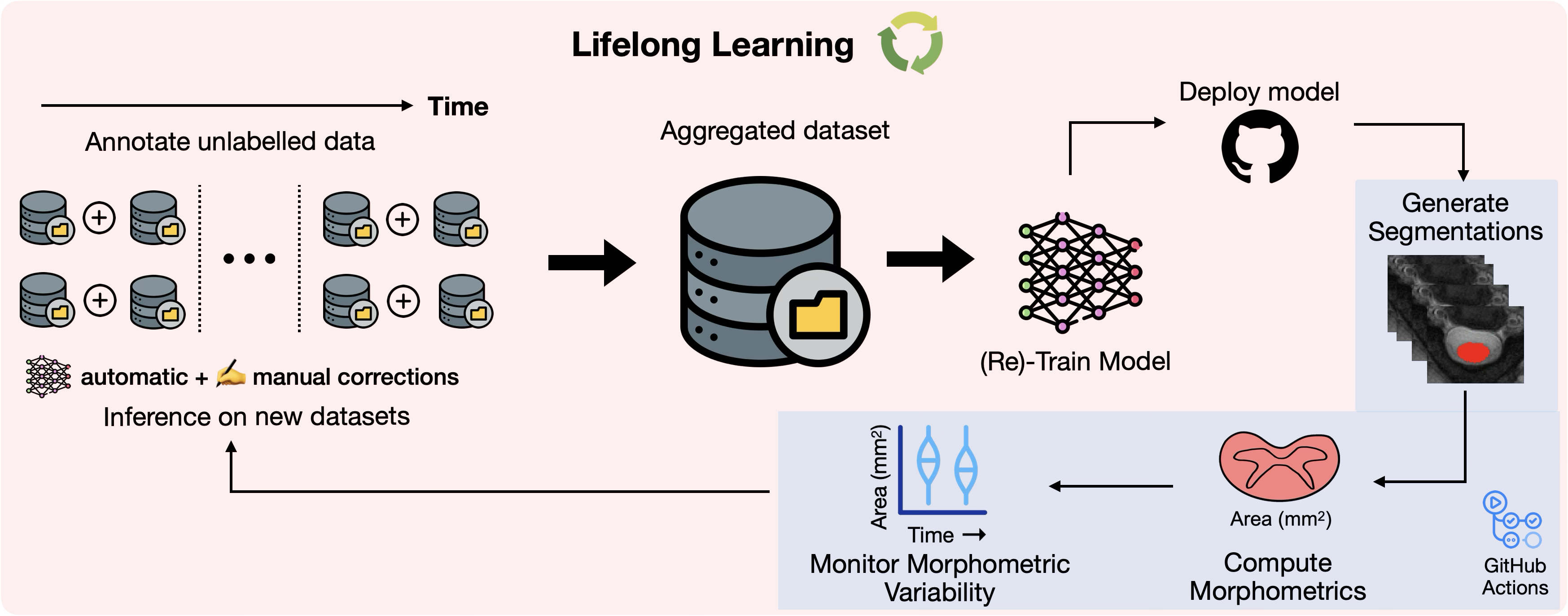
 Monitoring morphometric drift in lifelong learning segmentation of the spinal cordEnamundram Naga Karthik, Sandrine Bédard, Jan Valošek, and 53 more authorsarXiv, 2025
Monitoring morphometric drift in lifelong learning segmentation of the spinal cordEnamundram Naga Karthik, Sandrine Bédard, Jan Valošek, and 53 more authorsarXiv, 2025Morphometric measures derived from spinal cord segmentations can serve as diagnostic and prognostic biomarkers in neurological diseases and injuries affecting the spinal cord. While robust, automatic segmentation methods to a wide variety of contrasts and pathologies have been developed over the past few years, whether their predictions are stable as the model is updated using new datasets has not been assessed. This is particularly important for deriving normative values from healthy participants. In this study, we present a spinal cord segmentation model trained on a multisite (n=75) dataset, including 9 different MRI contrasts and several spinal cord pathologies. We also introduce a lifelong learning framework to automatically monitor the morphometric drift as the model is updated using additional datasets. The framework is triggered by an automatic GitHub Actions workflow every time a new model is created, recording the morphometric values derived from the model’s predictions over time. As a real-world application of the proposed framework, we employed the spinal cord segmentation model to update a recently-introduced normative database of healthy participants containing commonly used measures of spinal cord morphometry. Results showed that: (i) our model outperforms previous versions and pathology-specific models on challenging lumbar spinal cord cases, achieving an average Dice score of 0.95 ± 0.03; (ii) the automatic workflow for monitoring morphometric drift provides a quick feedback loop for developing future segmentation models; and (iii) the scaling factor required to update the database of morphometric measures is nearly constant among slices across the given vertebral levels, showing minimum drift between the current and previous versions of the model monitored by the framework. The model is freely available in Spinal Cord Toolbox v7.0.
@article{karthik2025monitoringmorphometricdriftlifelong, title = {Monitoring morphometric drift in lifelong learning segmentation of the spinal cord}, author = {Karthik, Enamundram Naga and Bédard, Sandrine and Valošek, Jan and Aigner, Christoph S. and Bannier, Elise and Bednařík, Josef and Callot, Virginie and Combes, Anna and Curt, Armin and David, Gergely and Eippert, Falk and Farner, Lynn and Fehlings, Michael G and Freund, Patrick and Granberg, Tobias and Granziera, Cristina and Group, RHSCIR Network Imaging and Horn, Ulrike and Horák, Tomáš and Humphreys, Suzanne and Hupp, Markus and Kerbrat, Anne and Kinany, Nawal and Kolind, Shannon and Kudlička, Petr and Lebret, Anna and Lee, Lisa Eunyoung and Mainero, Caterina and Martin, Allan R. and McGrath, Megan and Nair, Govind and O'Grady, Kristin P. and Oh, Jiwon and Ouellette, Russell and Pfender, Nikolai and Pfyffer, Dario and Pradat, Pierre-François and Prat, Alexandre and Pravatà, Emanuele and Reich, Daniel S. and Ricchi, Ilaria and Rotem-Kohavi, Naama and Schading-Sassenhausen, Simon and Seif, Maryam and Smith, Andrew and Smith, Seth A and Sweeney, Grace and Tam, Roger and Traboulsee, Anthony and Treaba, Constantina Andrada and Tsagkas, Charidimos and Vavasour, Zachary and Ville, Dimitri Van De and II, Kenneth Arnold Weber and Chandar, Sarath and Cohen-Adad, Julien}, year = {2025}, eprint = {2505.01364}, archiveprefix = {arXiv}, primaryclass = {cs.CV}, url = {https://arxiv.org/abs/2505.01364}, journal = {arXiv}, } - Imaging Neuroscience
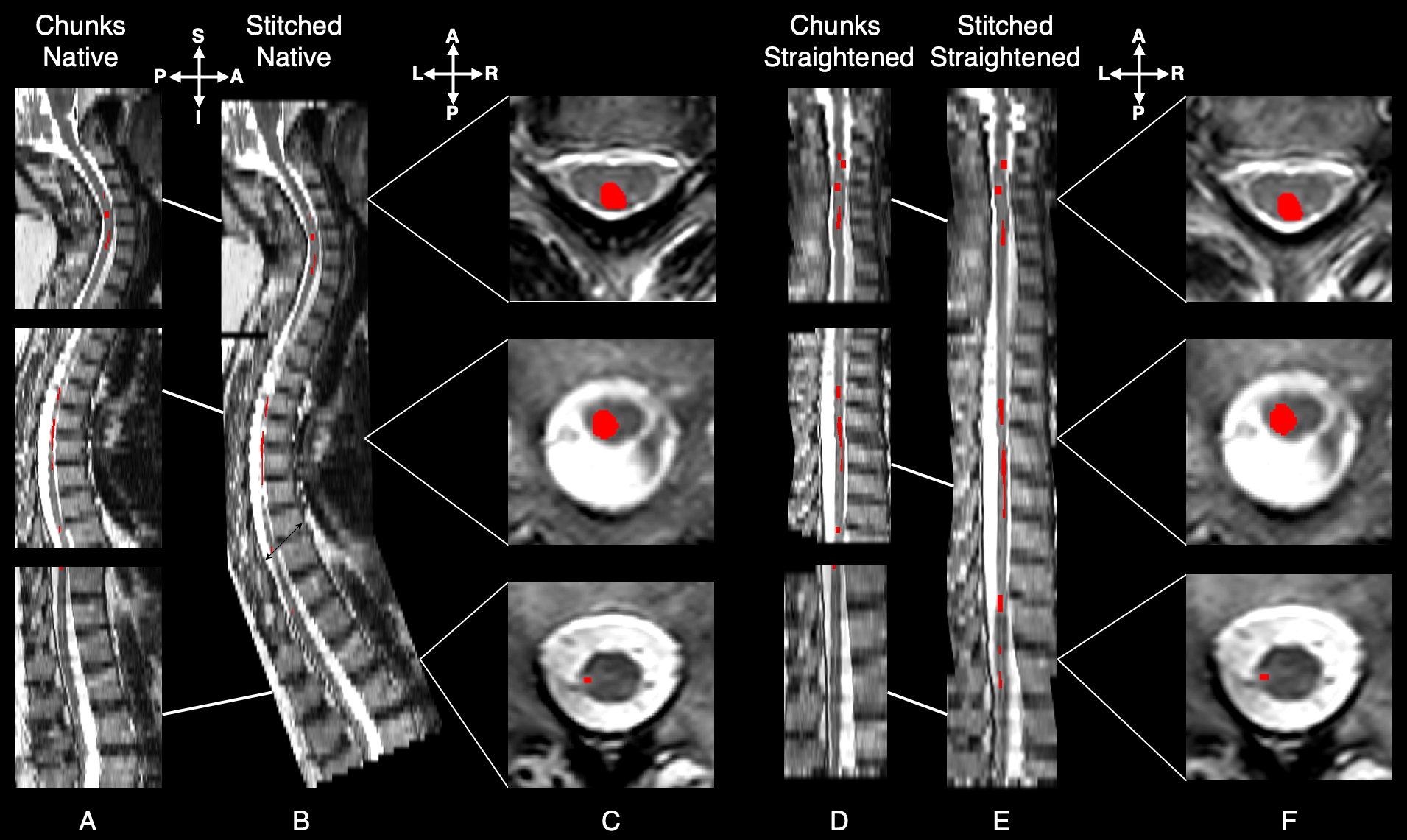 Automatic segmentation of spinal cord lesions in MS: A robust tool for axial T2-weighted MRI scansEnamundram Naga Karthik*, Julian McGinnis*, Ricarda Wurm, and 17 more authorsImaging Neuroscience, Jun 2025*shared first authorship
Automatic segmentation of spinal cord lesions in MS: A robust tool for axial T2-weighted MRI scansEnamundram Naga Karthik*, Julian McGinnis*, Ricarda Wurm, and 17 more authorsImaging Neuroscience, Jun 2025*shared first authorshipDeep learning models have achieved remarkable success in segmenting brain white matter lesions in multiple sclerosis (MS), becoming integral to both research and clinical workflows. While brain lesions have gained significant attention in MS research, the involvement of spinal cord lesions in MS is relatively understudied. This is largely owing to the variability in spinal cord magnetic resonance imaging (MRI) acquisition protocols, high individual anatomical differences, the complex morphology and size of spinal cord lesions, and lastly, the scarcity of labeled datasets required to develop robust segmentation tools. As a result, automatic segmentation of spinal cord MS lesions remains a significant challenge. Although some segmentation tools exist for spinal cord lesions, most have been developed using sagittal T2-weighted (T2w) sequences primarily focusing on cervical spines. With the growing importance of spinal cord imaging in MS, axial T2w scans are becoming increasingly relevant due to their superior sensitivity in detecting lesions compared to sagittal acquisition protocols. However, most existing segmentation methods struggle to effectively generalize to axial sequences due to differences in image characteristics caused by the highly anisotropic spinal cord scans. To address these challenges, we developed a robust, open-source lesion segmentation tool tailored specifically for axial T2w scans covering the whole spinal cord. We investigated key factors influencing lesion segmentation, including the impact of stitching together individually acquired spinal regions, straightening the spinal cord, and comparing the effectiveness of 2D and 3D convolutional neural networks (CNNs). Drawing on these insights, we trained a multi-center model using an extensive dataset of 582 MS patients, resulting in a dataset comprising an entirety of 2,167 scans. We empirically evaluated the model’s segmentation performance across various spinal segments for lesions with varying sizes. Our model significantly outperforms the current state-of-the-art methods, providing consistent segmentation across cervical, thoracic, and lumbar regions. To support the broader research community, we integrate our model into the widely-used Spinal Cord Toolbox (v7.0 and above), making it accessible via the command sct_deepseg lesion_ms_axial_t2 -i <path-to-image.nii.gz>.
@article{karthik2025automatic, author = {Naga Karthik, Enamundram and McGinnis, Julian and Wurm, Ricarda and Ruehling, Sebastian and Graf, Robert and Valosek, Jan and Benveniste, Pierre-Louis and Lauerer, Markus and Talbott, Jason and Bakshi, Rohit and Tauhid, Shahamat and Shepherd, Timothy and Berthele, Achim and Zimmer, Claus and Hemmer, Bernhard and Rueckert, Daniel and Wiestler, Benedikt and Kirschke, Jan S. and Cohen-Adad, Julien and Mühlau, Mark}, title = {Automatic segmentation of spinal cord lesions in MS: A robust tool for axial T2-weighted MRI scans}, journal = {Imaging Neuroscience}, volume = {3}, pages = {IMAG.a.45}, year = {2025}, month = jun, issn = {2837-6056}, doi = {10.1162/IMAG.a.45}, url = {https://doi.org/10.1162/IMAG.a.45}, note = {*shared first authorship}, } - MedIA
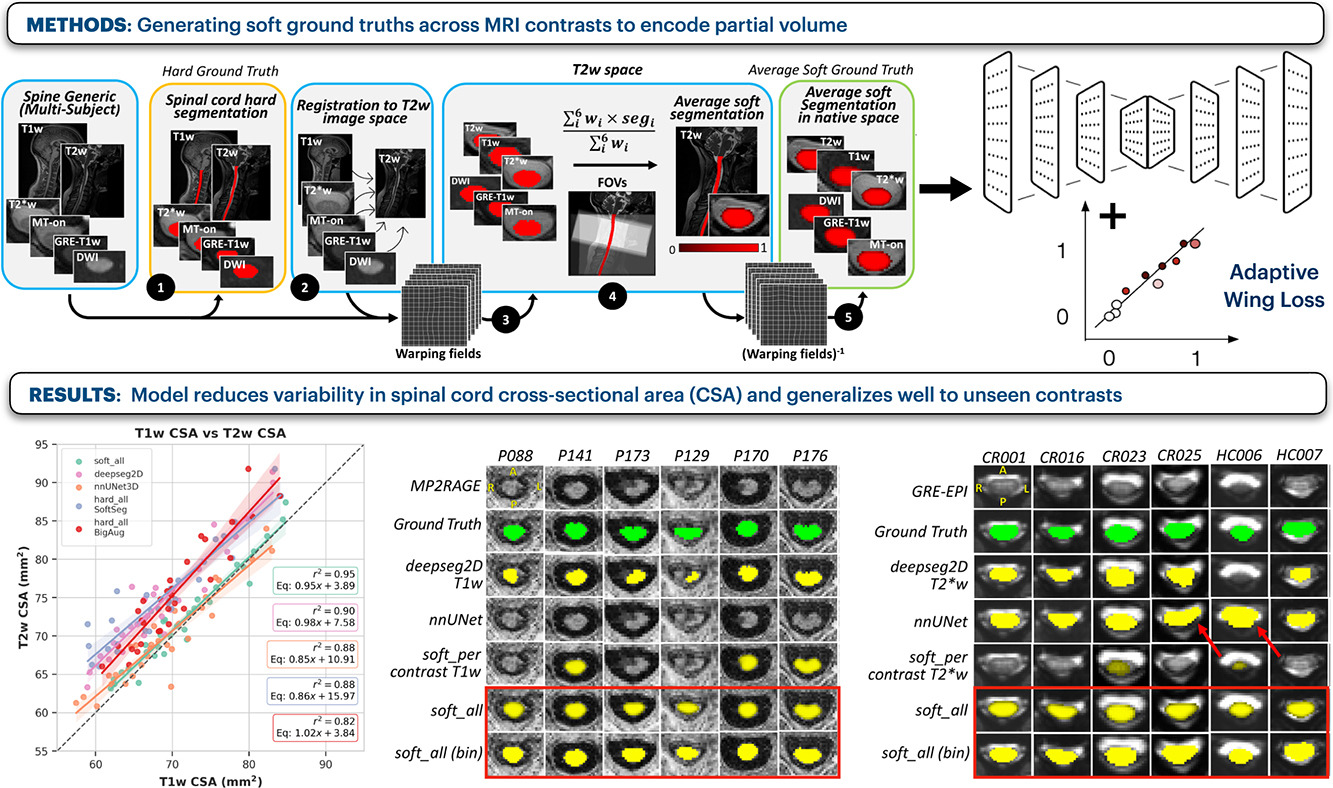 Towards contrast-agnostic soft segmentation of the spinal cordSandrine Bédard*, Enamundram Naga Karthik*, Charidimos Tsagkas, and 5 more authorsMedical Image Analysis, Jun 2025*shared first authorship
Towards contrast-agnostic soft segmentation of the spinal cordSandrine Bédard*, Enamundram Naga Karthik*, Charidimos Tsagkas, and 5 more authorsMedical Image Analysis, Jun 2025*shared first authorshipSpinal cord segmentation is clinically relevant and is notably used to compute spinal cord cross-sectional area (CSA) for the diagnosis and monitoring of cord compression or neurodegenerative diseases such as multiple sclerosis. While several semi and automatic methods exist, one key limitation remains: the segmentation depends on the MRI contrast, resulting in different CSA across contrasts. This is partly due to the varying appearance of the boundary between the spinal cord and the cerebrospinal fluid that depends on the sequence and acquisition parameters. This contrast-sensitive CSA adds variability in multi-center studies where protocols can vary, reducing the sensitivity to detect subtle atrophies. Moreover, existing methods enhance the CSA variability by training one model per contrast, while also producing binary masks that do not account for partial volume effects. In this work, we present a deep learning-based method that produces soft segmentations of the spinal cord that are stable across MRI contrasts. Using the Spine Generic Public Database of healthy participants (n=267; contrasts=6), we first generated participant-wise soft ground truth (GT) by averaging the binary segmentations across all 6 contrasts. These soft GT, along with aggressive data augmentation and a regression-based loss function, were then used to train a U-Net model for spinal cord segmentation. We evaluated our model against state-of-the-art methods and performed ablation studies involving different GT mask types, loss functions, contrast-specific models and domain generalization methods. Our results show that using the soft average segmentations along with a regression loss function reduces CSA variability (p<0.05, Wilcoxon signed-rank test). The proposed spinal cord segmentation model generalizes better than the state-of-the-art contrast-specific methods amongst unseen datasets, vendors, contrasts, and pathologies (compression, lesions), while accounting for partial volume effects. Our model is integrated into the Spinal Cord Toolbox (v6.2 and higher).
@article{BEDARD2025103473, title = {Towards contrast-agnostic soft segmentation of the spinal cord}, journal = {Medical Image Analysis}, volume = {101}, pages = {103473}, year = {2025}, issn = {1361-8415}, doi = {https://doi.org/10.1016/j.media.2025.103473}, url = {https://www.sciencedirect.com/science/article/pii/S1361841525000210}, author = {Bédard, Sandrine and Naga Karthik, Enamundram and Tsagkas, Charidimos and Pravatà, Emanuele and Granziera, Cristina and Smith, Andrew and {Weber II}, Kenneth Arnold and Cohen-Adad, Julien}, keywords = {Spinal cord, MRI, Contrasts, Segmentation, Deep learning, Soft labels, Partial volume effect}, note = {*shared first authorship}, } - Radiology: AI
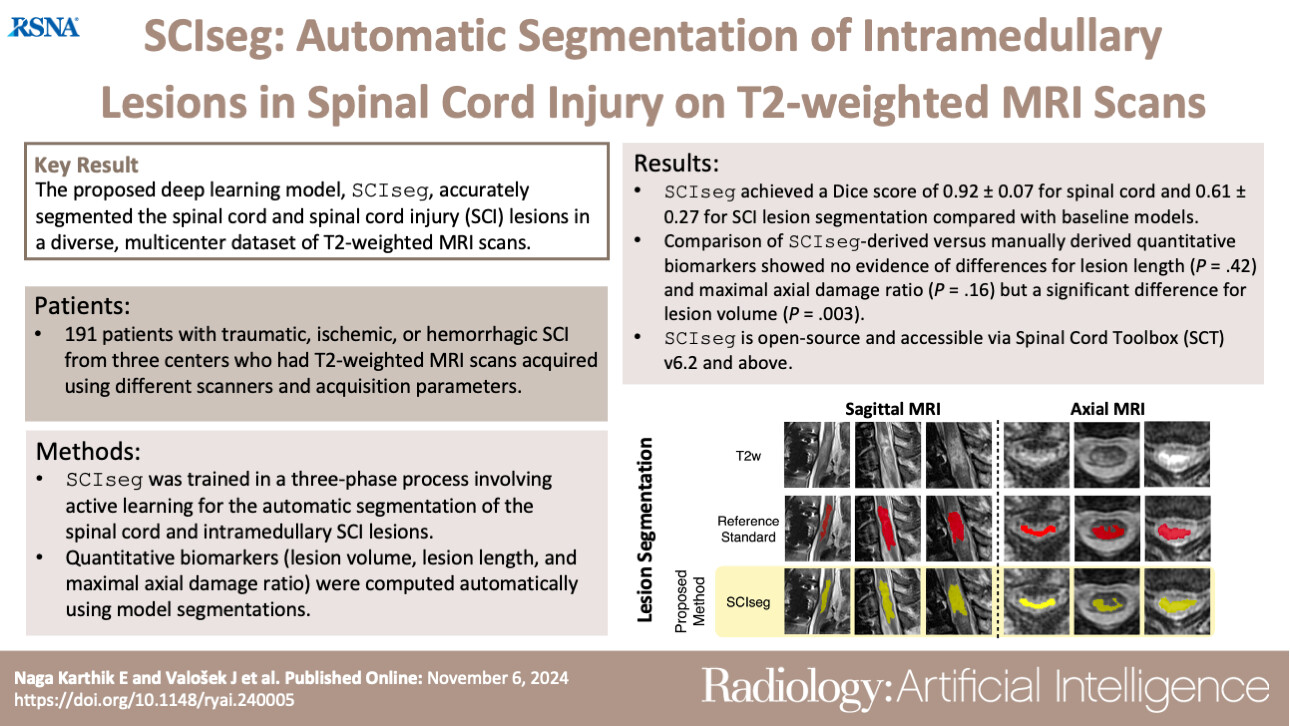 SCIseg: Automatic Segmentation of Intramedullary Lesions in Spinal Cord Injury on T2-weighted MRI ScansEnamundram Naga Karthik*, Jan Valošek*, Andrew C. Smith, and 6 more authorsRadiology: Artificial Intelligence, Jun 2025*shared first authorship
SCIseg: Automatic Segmentation of Intramedullary Lesions in Spinal Cord Injury on T2-weighted MRI ScansEnamundram Naga Karthik*, Jan Valošek*, Andrew C. Smith, and 6 more authorsRadiology: Artificial Intelligence, Jun 2025*shared first authorshipPurpose: To develop a deep learning tool for the automatic segmentation of the spinal cord and intramedullary lesions in spinal cord injury (SCI) on T2-weighted MRI scans. Materials and Methods: This retrospective study included MRI data acquired between July 2002 and February 2023. The data consisted of T2-weighted MRI scans acquired using different scanner manufacturers with various image resolutions (isotropic and anisotropic) and orientations (axial and sagittal). Patients had different lesion etiologies (traumatic, ischemic, and hemorrhagic) and lesion locations across the cervical, thoracic, and lumbar spine. A deep learning model, SCIseg (which is open source and accessible through the Spinal Cord Toolbox, version 6.2 and above), was trained in a three-phase process involving active learning for the automatic segmentation of intramedullary SCI lesions and the spinal cord. The segmentations from the proposed model were visually and quantitatively compared with those from three other open-source methods (PropSeg, DeepSeg, and contrast-agnostic, all part of the Spinal Cord Toolbox). The Wilcoxon signed rank test was used to compare quantitative MRI biomarkers of SCI (lesion volume, lesion length, and maximal axial damage ratio) derived from the manual reference standard lesion masks and biomarkers obtained automatically with SCIseg segmentations. Results: The study included 191 patients with SCI (mean age, 48.1 years ± 17.9 [SD]; 142 [74%] male patients). SCIseg achieved a mean Dice score of 0.92 ± 0.07 and 0.61 ± 0.27 for spinal cord and SCI lesion segmentation, respectively. There was no evidence of a difference between lesion length (P = .42) and maximal axial damage ratio (P = .16) computed from manually annotated lesions and the lesion segmentations obtained using SCIseg. Conclusion: SCIseg accurately segmented intramedullary lesions on a diverse dataset of T2-weighted MRI scans and automatically extracted clinically relevant lesion characteristics. Keywords: Spinal Cord, Trauma, Segmentation, MR Imaging, Supervised Learning, Convolutional Neural Network (CNN) Published under a CC BY 4.0 license.
@article{doi:10.1148/ryai.240005, author = {Naga Karthik, Enamundram and Valo\v{s}ek, Jan and Smith, Andrew C. and Pfyffer, Dario and Schading-Sassenhausen, Simon and Farner, Lynn and Weber, Kenneth A. and Freund, Patrick and Cohen-Adad, Julien}, title = {SCIseg: Automatic Segmentation of Intramedullary Lesions in Spinal Cord Injury on T2-weighted MRI Scans}, journal = {Radiology: Artificial Intelligence}, volume = {7}, number = {1}, pages = {e240005}, year = {2025}, note = {*shared first authorship}, }
2024
- MIDL ShortContrast-agnostic Spinal Cord Segmentation: A Comparative Study of ConvNets and Vision TransformersEnamundram Naga Karthik, Sandrine Bedard, Jan Valosek, and 2 more authorsIn Medical Imaging with Deep Learning, Jun 2024
The cross-sectional area (CSA) of the spinal cord (SC) computed from its segmentation is a relevant clinical biomarker for the diagnosis and monitoring of cord compression and atrophy. One key limitation of existing automatic methods is that their SC segmentations depend on the MRI contrast, resulting in different CSA across contrasts. Furthermore, these methods rely on CNNs, leaving a gap in the literature for exploring the performance of modern deep learning (DL) architectures. In this study, we extend our recent work \citeBdard2023TowardsCS by evaluating the contrast-agnostic SC segmentation capabilities of different classes of DL architectures, namely, ConvNeXt, vision transformers (ViTs), and hierarchical ViTs. We compared 7 different DL models using the open-source \textitSpine Generic Database of healthy participants () consisting of 6 MRI contrasts per participant. Given a fixed dataset size, our results show that CNNs produce robust SC segmentations across contrasts, followed by ConvNeXt, and hierarchical ViTs. This suggests that: (i) inductive biases such as learning hierarchical feature reprensentations via pooling (common in CNNs) are crucial for good performance on SC segmentation, and (ii) hierarchical ViTs that incorporate several CNN-based priors can perform similarly to pure CNN-based models.
@inproceedings{karthik2024contrastagnostic, title = {Contrast-agnostic Spinal Cord Segmentation: A Comparative Study of ConvNets and Vision Transformers}, author = {Karthik, Enamundram Naga and Bedard, Sandrine and Valosek, Jan and Chandar, Sarath and Cohen-Adad, Julien}, booktitle = {Medical Imaging with Deep Learning}, year = {2024}, url = {https://openreview.net/forum?id=n6D25aqdV3}, }
2023
- IEEE OJEMB
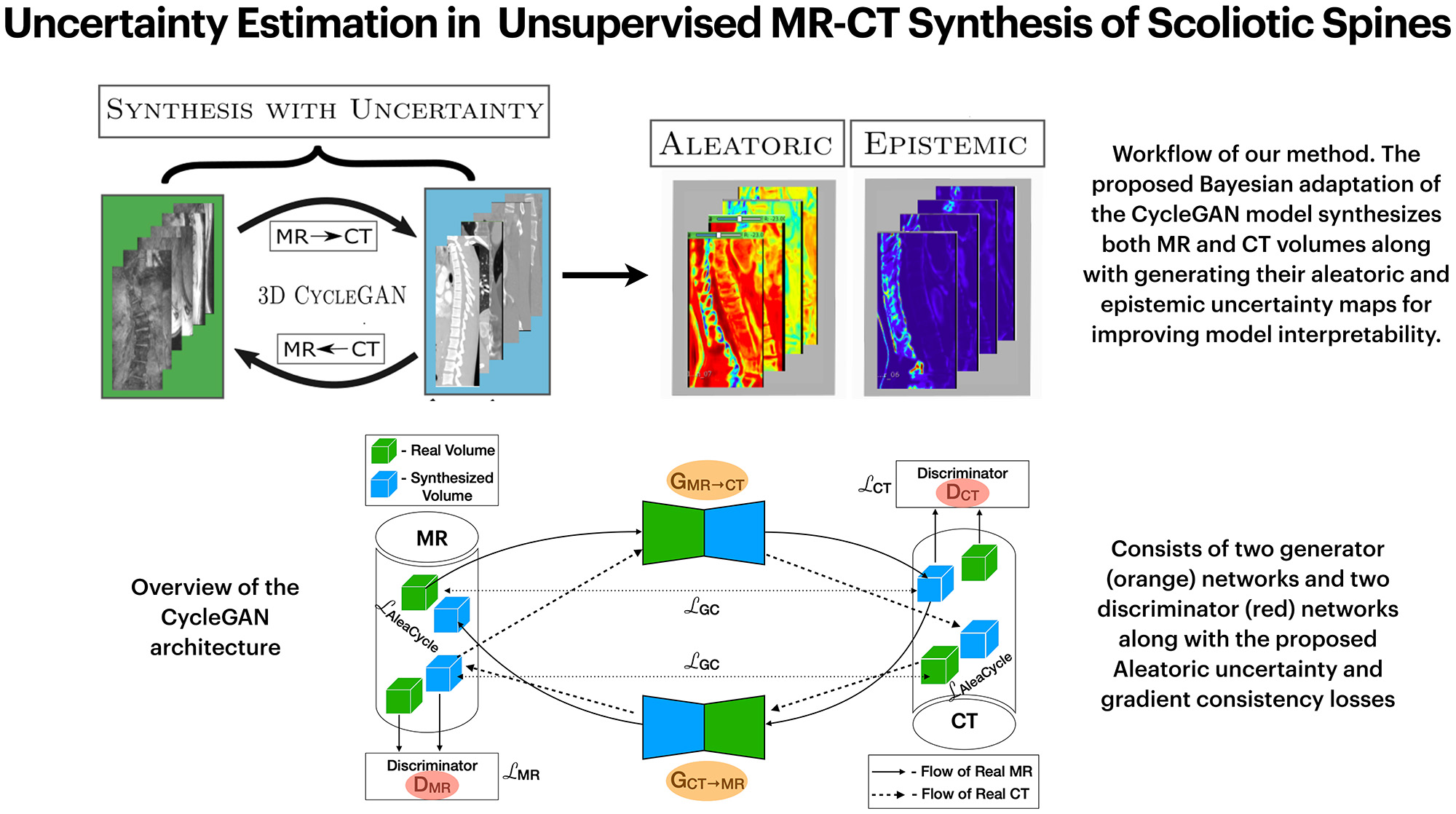 Uncertainty Estimation in Unsupervised MR-CT Synthesis of Scoliotic SpinesEnamundram Naga Karthik, Farida Cheriet, and Catherine LaporteIEEE Open Journal of Engineering in Medicine and Biology, Jun 2023
Uncertainty Estimation in Unsupervised MR-CT Synthesis of Scoliotic SpinesEnamundram Naga Karthik, Farida Cheriet, and Catherine LaporteIEEE Open Journal of Engineering in Medicine and Biology, Jun 2023Uncertainty estimations through approximate Bayesian inference provide interesting insights to deep neural networks’ behavior. In unsupervised learning tasks, where expert labels are unavailable, it becomes ever more important to critique the model through uncertainties. This paper presents a proof-of-concept for generalizing the aleatoric and epistemic uncertainties in unsupervised MR-CT synthesis of scoliotic spines. A novel adaptation of the cycle-consistency constraint in CycleGAN is proposed such that the model predicts the aleatoric uncertainty maps in addition to the standard volume-to-volume translation between Magnetic Resonance (MR) and Computed Tomography (CT) data. Ablation experiments were performed to understand uncertainty estimation as an implicit regularizer and a measure of the model’s confidence. The aleatoric uncertainty helps in distinguishing between the bone and soft-tissue regions in CT and MR data during translation, while the epistemic uncertainty provides interpretable information to the user for downstream tasks.
@article{10086579, author = {Naga Karthik, Enamundram and Cheriet, Farida and Laporte, Catherine}, journal = {IEEE Open Journal of Engineering in Medicine and Biology}, title = {Uncertainty Estimation in Unsupervised MR-CT Synthesis of Scoliotic Spines}, year = {2023}, volume = {}, number = {}, pages = {1-7}, doi = {https://doi.org/10.1109/OJEMB.2023.3262965}, }
2022
- MELBA
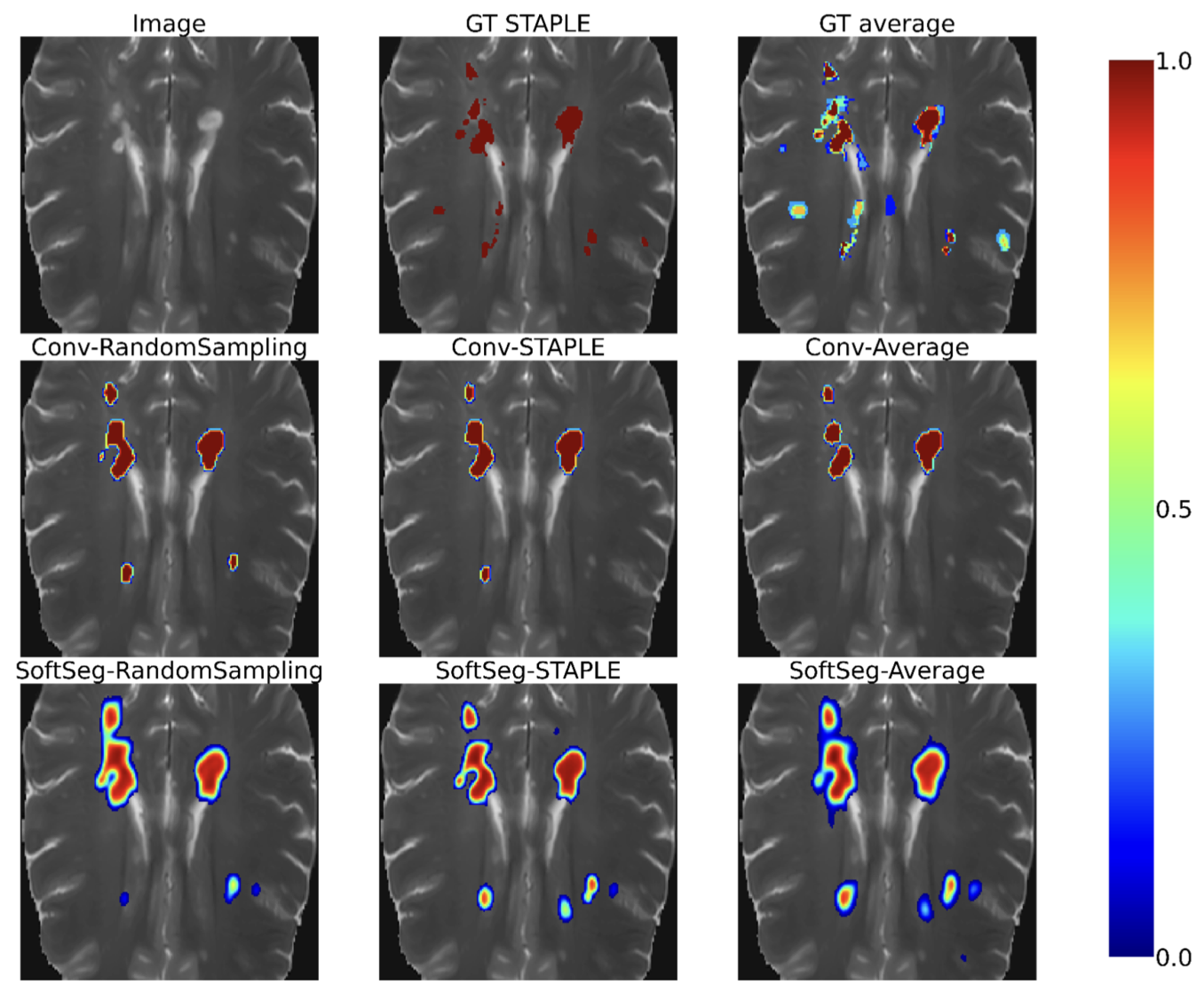 Label fusion and training methods for reliable representation of inter-rater uncertaintyAndreanne Lemay, Charley Gros, Enamundram Naga Karthik, and 1 more authorMachine Learning for Biomedical Imaging, Jun 2022
Label fusion and training methods for reliable representation of inter-rater uncertaintyAndreanne Lemay, Charley Gros, Enamundram Naga Karthik, and 1 more authorMachine Learning for Biomedical Imaging, Jun 2022Medical tasks are prone to inter-rater variability due to multiple factors such as image quality, professional experience and training, or guideline clarity. Training deep learning networks with annotations from multiple raters is a common practice that mitigates the model’s bias towards a single expert. Reliable models generating calibrated outputs and reflecting the inter-rater disagreement are key to the integration of artificial intelligence in clinical practice. Various methods exist to take into account different expert labels. We focus on comparing three label fusion methods: STAPLE, average of the rater’s segmentation, and random sampling of each rater’s segmentation during training. Each label fusion method is studied using both the conventional training framework and the recently published SoftSeg framework that limits information loss by treating the segmentation task as a regression. Our results, across 10 data splittings on two public datasets (spinal cord gray matter challenge, and multiple sclerosis brain lesion segmentation), indicate that SoftSeg models, regardless of the ground truth fusion method, had better calibration and preservation of the inter-rater rater variability compared with their conventional counterparts without impacting the segmentation performance. Conventional models, i.e., trained with a Dice loss, with binary inputs, and sigmoid/softmax final activate, were overconfident and underestimated the uncertainty associated with inter-rater variability. Conversely, fusing labels by averaging with the SoftSeg framework led to underconfident outputs and overestimation of the rater disagreement. In terms of segmentation performance, the best label fusion method was different for the two datasets studied, indicating this parameter might be task-dependent. However, SoftSeg had segmentation performance systematically superior or equal to the conventionally trained models and had the best calibration and preservation of the inter-rater variability. SoftSeg has a low computational cost and performed similarly in terms of uncertainty to ensembles which require multiple models and forward passes. Our code is available at https://ivadomed.org.
@article{melba:2022:031:lemay, title = {Label fusion and training methods for reliable representation of inter-rater uncertainty}, author = {Lemay, Andreanne and Gros, Charley and Naga Karthik, Enamundram and Cohen-Adad, Julien}, journal = {Machine Learning for Biomedical Imaging}, volume = {1}, issue = {January 2023 issue}, year = {2022}, pages = {1--27}, issn = {2766-905X}, doi = {https://doi.org/10.59275/j.melba.2022-db5c}, url = {https://melba-journal.org/2022:031}, } - MedNeurIPSSegmentation of Multiple Sclerosis Lesion across Hospitals: Learn Continually or Train from Scratch?Enamundram Naga Karthik, Anne Kerbrat, Pierre Labauge, and 8 more authorsMedNeurIPS: Medical Imaging Meets NeurIPS Workshop, Jun 2022
@article{nagakarthik2022Segmentation, title = {Segmentation of Multiple Sclerosis Lesion across Hospitals: Learn Continually or Train from Scratch?}, author = {Naga Karthik, Enamundram and Kerbrat, Anne and Labauge, Pierre and Granberg, Tobias and Talbott, Jason and Reich, Daniel S and Filippi, Massimo and Bakshi, Rohit and Callot, Virginie and Chandar, Sarath and Cohen-Adad, Julien}, journal = {MedNeurIPS: Medical Imaging Meets NeurIPS Workshop}, year = {2022}, url = {https://arxiv.org/pdf/2210.15091.pdf}, }
2021
- SPIEThree-dimensional segmentation of the scoliotic spine from MRI using unsupervised volume-based MR-CT synthesisEnamundram M. V. Naga Karthik, Catherine Laporte, and Farida CherietIn Medical Imaging 2021: Image Processing, Jun 2021
@inproceedings{10.1117/12.2580677, author = {Karthik, Enamundram M. V. Naga and Laporte, Catherine and Cheriet, Farida}, title = {{Three-dimensional segmentation of the scoliotic spine from MRI using unsupervised volume-based MR-CT synthesis}}, volume = {11596}, booktitle = {Medical Imaging 2021: Image Processing}, editor = {Išgum, Ivana and Landman, Bennett A.}, organization = {International Society for Optics and Photonics}, publisher = {SPIE}, pages = {115961H}, keywords = {Vertebrae segmentation , Scoliosis , Cross-modality synthesis, Volume translation , 3D CycleGAN}, year = {2021}, doi = {10.1117/12.2580677}, url = {https://doi.org/10.1117/12.2580677}, }
2020
- JASAAutomatic tongue surface extraction from three-dimensional ultrasound vocal tract imagesEnamundram M. V. Naga Karthik, Elham Karimi, Steven M. Lulich, and 1 more authorThe Journal of the Acoustical Society of America, Jun 2020
@article{doi:10.1121/10.0000891, author = {Naga Karthik, Enamundram M. V. and Karimi, Elham and Lulich, Steven M. and Laporte, Catherine}, title = {Automatic tongue surface extraction from three-dimensional ultrasound vocal tract images}, journal = {The Journal of the Acoustical Society of America}, volume = {147}, number = {3}, pages = {1623-1633}, year = {2020}, doi = {10.1121/10.0000891}, url = {https://doi.org/10.1121/10.0000891}, eprint = {https://doi.org/10.1121/10.0000891}, }
1935
- Can Quantum-Mechanical Description of Physical Reality Be Considered Complete?A. Einstein*†, B. Podolsky*, and N. Rosen*Phys. Rev., New Jersey. More Information can be found here , May 1935
In a complete theory there is an element corresponding to each element of reality. A sufficient condition for the reality of a physical quantity is the possibility of predicting it with certainty, without disturbing the system. In quantum mechanics in the case of two physical quantities described by non-commuting operators, the knowledge of one precludes the knowledge of the other. Then either (1) the description of reality given by the wave function in quantum mechanics is not complete or (2) these two quantities cannot have simultaneous reality. Consideration of the problem of making predictions concerning a system on the basis of measurements made on another system that had previously interacted with it leads to the result that if (1) is false then (2) is also false. One is thus led to conclude that the description of reality as given by a wave function is not complete.